DV200 evaluation using RNA ScreenTape analysis
Probiotics are a class of active microorganisms that are beneficial to the host by colonizing the human body and changing the composition of the flora in a certain part of the host. By regulating the immune function of the host mucosa and system or by regulating the balance of intestinal flora, promoting nutrient absorption and maintaining intestinal health, resulting in the production of single microorganisms or mixed microorganisms with well-defined composition that are beneficial to health. Probiotics,Probiotic Efficacy,Probiotic vitamins,Probiotic Health Supplements YT(Xi'an) Biochem Co., Ltd. , https://www.ytlinkherb.com
Eva Graf
Agilent Technology Co., Ltd.
Waldbronn, Germany
Summary
The success of the RNA sequencing library preparation workflow depends to a large extent on the quality of the RNA starting material. The RNA Complete Value Equivalent (RINe) obtained using the Agilent RNA ScreenTape assay is a reliable and reproducible indicator of the integrity of a sample for standard RNA sequencing. Due to RNA degradation and limited sample volume, extraction of RNA samples from formalin-fixed paraffin-embedded (FFPE) tissue for library preparation is challenging, so a tailored approach to FFPE RNA library preparation is recommended. Fragment size distribution of FFPE RNA samples has a significant impact on library yield, which can be described as a percentage of RNA fragments greater than 200 nucleotides, expressed as DV 200 quality indicators [1] . The Agilent RNA ScreenTape and Agilent High Sensitivity RNA ScreenTape assays facilitate highly reproducible DV 200 assessment of degraded RNA samples extracted from FFPE tissue. Agilent TapeStation analysis software enables easy evaluation and processing of DV 200 data through regional analysis, automatically performing repeated DV 200 analysis.
Technical details
An Agilent 4200 TapeStation system with Agilent TapeStation analysis software (version A.02.02) was used. RNA ScreenTape (p/n 5067-5576) and reagents (part numbers 5067-5577 and 5067-5578), as well as Agilent's high sensitivity RNA ScreenTape (part number 5067-5579) and reagents (part numbers 5067-5580 and 5067-5581) Purchased from Agilent Technologies. All analyses were performed according to the manufacturer's protocol and guidelines unless otherwise stated. All samples were analyzed using the ScreenTape assay, and each strip contained a gradient molecular weight internal standard loading. RNA samples extracted from FFPE tissue were provided by the National Cancer Center (NCT, Heidelberg, Germany), which was organized in accordance with the organization's library and approved by the Ethics Committee of the University of Heidelberg.
data analysis
DV 200 represents the percentage of RNA fragments greater than 200 nucleotides in all RNA fragments. By defining a region greater than 200 nucleotides, DV 200 can be calculated using the TapeStation analysis software. The software offers a variety of locale options for a wide range of sample throughputs from low to high. For more information and a demo video on regional analysis, please consult the Agilent Information Center (AIC).
200 area of a single sample locale <br> defined as a single sample DV, click the Region, and then right-click to add the zone electrophoresis. The area boundaries can be individually adjusted by swiping left or right.
<br> locale data files if you want to DV 200 area defined as the default applied to all samples in a single data file, click the Region, and then clicking Region Settings, lower and upper limits input area (Figure 1) in the dialog box . To identify this area in the electropherogram, enter DV 200 as the area comment. To apply this area to all samples in the active file, select This File . 
Auto locale <br> To DV 200 area defined as the default for all repeat RNA FFPE sample analysis, click the Region, and then clicking Region Settings, lower and upper limits input area (Figure 1) in the dialog box. To identify this area in the electropherogram, enter
The DV 200 is used as a regional comment. To apply this DV 200 area to all files that are identical to the active file analysis method, select Assay Files .
Once the DV 200 region definition is complete, the value for each sample will be displayed in the % of Total column in the region table (Figure 2). You can export all the area data and generate a report by selecting the area table. Note that the concentration displayed in the area table, export file, and report of the TapeStation analysis software refers to the concentration of the defined area; the total sample concentration is shown in the sample table. 
Results and discussion
<br> three reproducible FFPE RNA samples were repeated five independent assay to verify the reproducibility DV 200 was obtained in RNA and RNA ScreenTape high sensitivity assay. Both methods show high reproducibility with a CV of less than 5% (Figure 3). 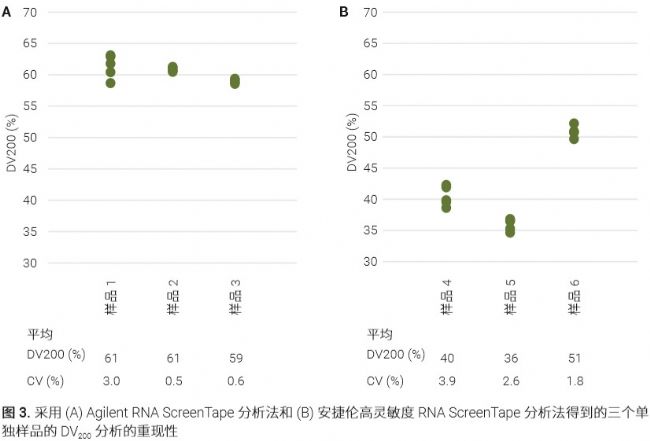
<br> concentration dependent RNA using RNA ScreenTape and high sensitivity analysis of three separate dilution series was FFPE RNA samples were analyzed to determine the consistency of the analysis 200 DV (Table 1). The DV 200 obtained by the RNA ScreenTape assay and the high-sensitivity RNA ScreenTape assay showed high accuracy with a CV value of less than 5%. 
As shown in Figure 4, a series of dilute solution analyses showed that the DV 200 evaluation was the most accurate when the measured concentration was above the lower limit of the specified analytical concentration range. 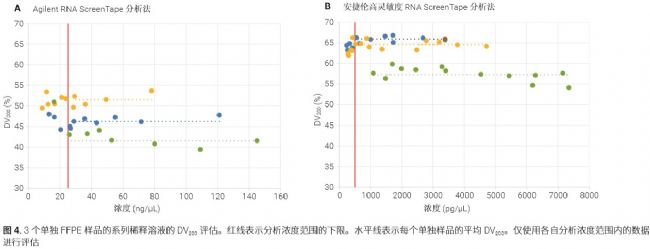
Severely degraded FFPE samples may move down to near the lower molecular weight internal standard and not completely separate from the lower internal standard, especially when the sample concentration is close to the upper limit of the specific quantitation range. In this case, the DV 200 results from the diluted sample are more accurate (see Figure 5).
Comparison of analytical methods <br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> The DV 200 obtained by RNA analysis ranged from 40% to 74%, while the results of analysis of the corresponding samples by high sensitivity RNA analysis were 101 ± 5% of these values. To verify the comparability of DV 200 measured by RNA and the high-sensitivity RNA ScreenTape assay, the values ​​obtained by the two methods are plotted together, as shown in Figure 6.
in conclusion
Agilent RNA and Agilent's high-sensitivity RNA ScreenTape assay, combined with the Agilent 4200 TapeStation system, make mass analysis of FFPE RNA samples quickly and easily. The Agilent TapeStation analysis software displays the DV 200 as a percentage of the total. The RINe algorithm does not affect the calculation of the DV 200 . DV 200 was calculated by RNA and high sensitivity RNA ScreenTape analysis with high reproducibility, and the results obtained by the two methods were highly consistent. For best performance, it is recommended to perform gradient molecular weight internal standard analysis with the sample. The DV 200 of the serial dilution solution is consistent over a specific analytical range. For highly degraded samples that are not effectively separated from the internal standard, they should be diluted to obtain the most accurate results.
references
1. Evaluating RNA Quality from FFPE Samples. Illumina Technical Note , publication number 470-2014-001. https:// technote- truseq-rna-access.pdf
For research use only. Not for diagnostic purposes.
Information, descriptions, and specifications in this publication are subject to change without notice.
© Agilent Technologies (China) Co., Ltd., 2017
September 28, 2017, China Publishing
5991-8355ENCN